Introduction
This protocol describes the approach used to stain formalin-fixed paraffin-embedded liver biopsy slides with a panel of metal-conjugated antibodies for subsequent analysis by Standard BioTool Hyperion imaging mass cytometry (IMC) as reported previously(1). The panel specifically focused on HBV, hepatic structural and immune markers as follows:-
- HBV antigens (HBsAg and HBcAg)
- Hepatocytes, biliary epithelial cells, endothelial cells, collagen and nuclei
- Innate and adaptive immune cells plus markers of memory, activation and function
Some tips to consider:
-
- Best to use recently cut slides with better preserved antigens for staining.
- Helpful to have a concurrent H&E staining to know the general biopsy characteristics.
- Antibodies that work in cell suspension mass cytometry do not necessarily work in IMC. Thus, it is best to test the antibodies with your samples in various dilutions (including pre-conjugated antibodies from Standard BioTool (previously called Fluidigm)).
- In designing the antibody panel, be mindful of potential mass cytometry spillover related to isotopic impurities and oxidation as well as abundance of a marker. This can be managed by reviewing isotope purity matrix (available from Standard BioTool) to avoid selecting isotopes with high spillover for highly expressed markers and by controls with single antibody staining to look for spillover into other channels. An R-based software from Bodenmiller lab can also be applied(2).
- Stained/fixed slides are generally stable for additional acquisition at a later time.
- Due to laser ablation involved in IMC, acquired tissue region on the slide cannot be used for further staining and analyses.
Helpful links for Analytic Workflow and other processes:
https://bodenmillergroup.github.io/IMCDataAnalysis/index.html
Materials and Reagents
Note: All water (including that used for EtOH dilutions, buffers etc.) should be 18.2 MΩ-cm purified.
- Antigen retrieval buffer: Tris-EDTA Buffer 10mM Tris Base, 1mM EDTA, 0.05% Tween 20 Solution, pH 9.0. To prepare 1L, dissolve 1.21 g Tris Base,37 g EDTA, 1000 ml water. If necessary, pH to pH 9.0 with HCl or NaOH. Add 0.5ml Tween 20. Store at RT.
- 10% BSA, for preparation of blocking solution and antibody cocktail. Prepare fresh each time, or dilute from frozen aliquots.
- 500 µM stock of iridium (Standard BioTool, SKU#201192B)
- Plastic Coplin jars, for dewaxing, rehydrating and rinsing tissue sections.
- A pressure cooker for antigen retrieval. We use the BioCare decloaking chamber (BioCare, SKU# DC2012)
- A humidified chamber. (We use slide storage boxes with wet paper towels laid across the bottom.)
- PAP Pen
Experimental Procedures
Staining Procedure
- Obtain tissue sections from FFPE blocks cut at ~3-8μm thick (We typically use 5μm.) mounted on charged slides.
Note: Stain the slides soon after the sections are mounted. If the slides are not to be stained immediately, store slides at -80℃ to slow epitope degradation and maximize staining quality. Slides should typically be stained within a few weeks, but the dependence of staining viability on this length of time will be dependent on the specific antibody clones in your antibody panel.
- Prepare the following washes in Coplin jars for dewaxing slides and rehydrating tissue sections:a. Xylene b. Xylene c. 100% ethanol d. 100% ethanol e. 95% ethanol f. 80% ethanol g. 70 % ethanol h. Water i. Water
- Preheat the antigen retrieval buffer in the pressure cooker to 95℃.
- Dewax the slides in a fume hood by sequential immersion in 2 baths of xylene for 15 minutes each.
- Rehydrate the sections by immersion in the Coplin jars containing 100-70% ethanol for 3 minutes each, then the two water washes for 1 minute each.
- Perform antigen retrieval by immersing the slides in the 95℃ antigen retrieval buffer. Pressure-cook for 30 minutes.
- Turn off the pressure cooker, remove the lid, and allow the slides to cool to RT.
- Rinse the slides by transferring them to a Coplin jar containing PBS.
- Remove the slides one by one and, without letting the slides dry (and deliberately allowing copious amounts of PBS to remain on the section). Encircle each tissue section with the PAP pen. Place each slide horizontally on the bench, and pipet PBS onto the surface of the tissue section if necessary.
- Prepare the blocking buffer by diluting the 10% BSA to 3% in PBS. Remove the PBS and add a blocking buffer. Incubate at RT for at least 1 hr, in the humidified chamber.
- Prepare your antibody cocktail at previously optimized concentrations in PBS, allowing room for adding 10% BSA to a final concentration of 0.5%.
- Remove the blocking buffer and add the antibody cocktail. Incubate the slides in the humidified chamber overnight at 4℃.
- Remove the antibody cocktail and stain DNA by incubating in 1.25 µM iridium in PBS. Incubate for 20 minutes.
- Wash slides in an excess of PBS in a Coplin jar for 5min.
- Wash slides again by sequential immersion in 2 baths of water for 2 minutes each.
- Air dry and store slides at RT until image acquisition.
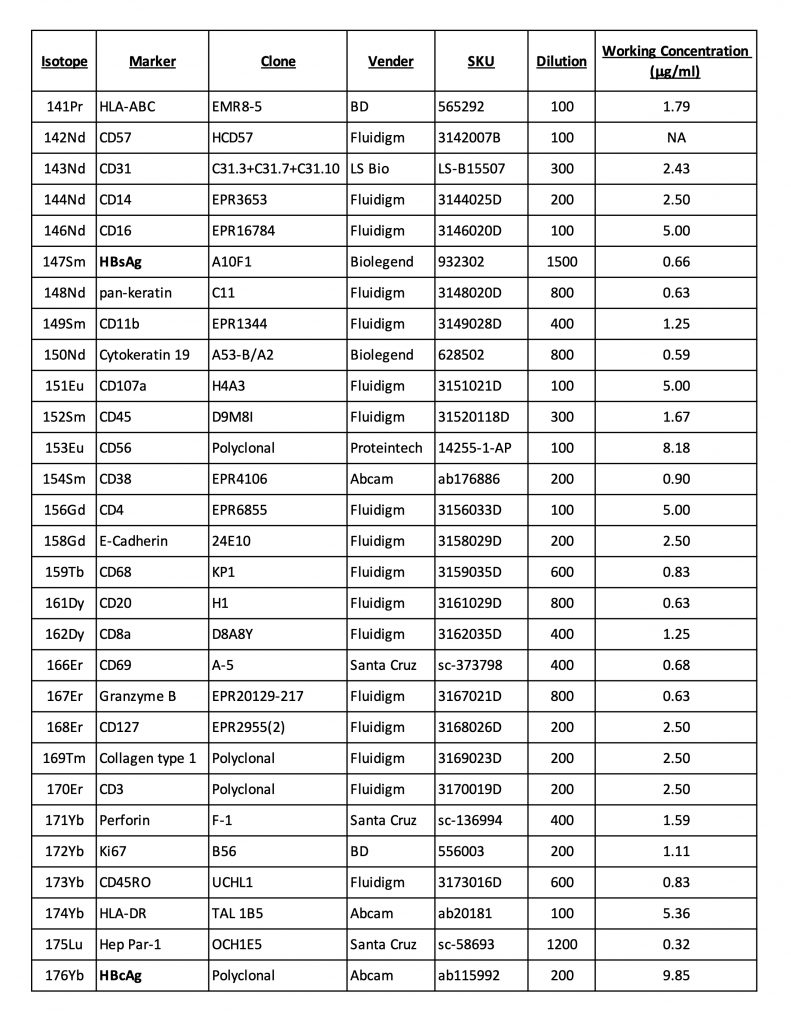
Antibody Panel
This sample panel consists of antibodies conjugated manually using Maxpar® X8 Antibody Labeling Kits (Standard BioTool) per the manufacturer’s recommendation and antibodies that are available in pre-conjugated forms, in addition to Iridium 191/193 for nuclear staining. This is meant only to be an example to be modified based on research interests and available antibodies.Image Acquisition by Hyperion IMC
- Select regions of interest (ROIs) in each slide/image based on bright-field images with the inclusion of both portal and lobular regions.
- Acquire an image as per the manufacturer’s standard operating procedures with a 200Hz laser frequency and a 1μm step increment.
- Note that 1mm2 ROI typically requires ~98 minutes to acquire, and the Hyperion can run overnight. This can be programmed in a way that the same slide can be acquired at multiple locations within the same tissue.
- Because liver biopsies come in various shapes and sizes, multiple adjacent ROIs may need to be acquired and combined to construct the final image.
- Note: 1.8-2mm2 total region acquired per liver biopsy sample provided statistically significant differences in immune cell frequency and portal expansion in our study.
Image Visualization
Images can be visualized individually or as overlays of multiple channels. Typically, it is helpful to combine structural elements (e.g. nuclei, collagen or epithelial markers, hepatocyte marker Hep Par-1) that provide some landmark information such as portal vs lobular region first, followed by other markers of interest (e.g. viral, immune).
Relevant software applications and links are shown below:
- https://imagej.nih.gov/ij/download.html
- HistoCAT++https://github.com/BodenmillerGroup/histoCAT/releases
- https://bodenmillergroup.github.io/IMCDataAnalysis/image-visualization.html
- MCD Viewer: https://www.standardbio.com/products/software
Image Analysis and Segmentation Strategies
- Reconstruct acquired images for all channels and multiple ROIs with preprocessing in ImageJ with a 3x3 pixel median filter to remove ‘salt-and-pepper' noise
- Establish a portal mask to distinguish between the portal and lobular region, as a binary image to be included along with the stained channels for analysis.
- Define cell segmentation strategy based on cells of interest and their characteristics in the tissue. For example, as immune cells can be tightly clustered in the portal region or highly inflamed tissue, we defined 1 micron beyond the nucleus as the cellular area. This may not be as suitable for larger cells (e.g. hepatocytes).
- Conduct cell segmentation using CellProfiler (3.0.0)
- Export CSV files from CellProfiler with the following information per segmented cell:
- Mean expression levels for every marker included in the antibody panel, and the binary portal mask.
- x/y coordinates to provide a 2-dimensional location within the acquired image.
- Convert expression values into z-scores for each image and normalize as an internal standardization step to minimize variations between ROIs acquired over time. This only works if there are clear positive and negative populations for each marker, in every ROI.
- CSV files exported from Cell Profiler or R can be:
- Converted to FCS files by FlowJo for analysis within FlowJo. Also, Manual cut-off values chosen in FlowJo can be exported for further analysis in R.
- Import into R for cluster analysis and visualization (e.g. PhenoGraph, tSNE, UMAP…)
References
- Traum D, Wang YJ, Schwarz KB, Schug J, Wong DK, Janssen H, Terrault NA, Khalili M, Wahed AS, Murray KF, Rosenthal P, Ling SC, Rodriguez-Baez N, Sterling RK, Lau DT, Block TM, Feldman MD, Furth EE, Lee WM, Kleiner DE, Lok AS, Kaestner KH, Chang KM. 2021. Highly multiplexed 2-dimensional imaging mass cytometry analysis of HBV-infected liver. JCI Insight 6.
- Chevrier S, Crowell HL, Zanotelli VRT, Engler S, Robinson MD, Bodenmiller B. 2018. Compensation of Signal Spillover in Suspension and Imaging Mass Cytometry. Cell Syst 6:612-620 e5.