Experimental Procedures
This method has previously been described in greater detail as a chapter in the book
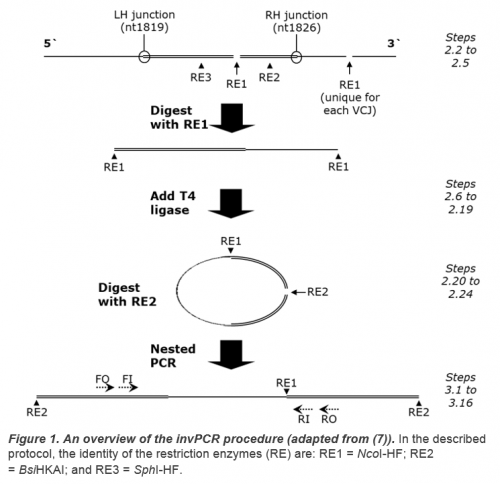
Methods in Molecular Biology: Hepatitis B Virus (7) and as an online open-access video in the Journal of Visualised Experiments (10). The procedure is summarized in Figure 1.
1A. Extraction of DNA (liver tissue from HBV-infected patients)
- Within a biosafety cabinet, use a sterile plastic Petri dish and a sterile scalpel blade to excise a ~5 mg fragment from a snap-frozen sample of biopsy or autopsy liver tissue.
- Place the tissue immediately in a 2 mL screw-cap Eppendorf tube with 400 μL of digestion solution (100 mM NaCl, 0.5% SDS, 50 mM Tris pH 7.5, 10 mM EDTA, 2 mg/mL proteinase K).
- Incubate in a thermomixer rotating at 3,000 rpm at 55°C for >2 hr (extendable to overnight) until no obvious pieces of liver parenchyma are visible. Some pieces of fibrotic tissue or fat may still be obvious.
- In a fume cupboard, add 400 μL of UltraPure Phenol and mix by inversion.
- Centrifuge at 14,000 g for 10 min and transfer the top aqueous layer to a clean 2 mL screw-cap Eppendorf tube, taking care to avoid any milky or cloudy interface.
- Repeat steps 4-5 twice with 400 μL of 25:24:1 UltraPure phenol:chloroform:isoamyl alcohol in the place of the phenol.
- Add 35 μL of 3 M sodium acetate pH 4.6 to the extracted DNA, followed by of 800 μL of AR grade ethanol.
- Incubate at -20°C overnight or at -80°C for 4 hr to precipitate the DNA.
- Pellet the precipitated DNA by centrifugation at 14,000 g.
- Wash the pellet with 1 mL of 70% ethanol.
- Repeat Step 10.
- Vacuum-dry for 20 min and redissolve in 50 μL of DNase-free water or 100 mM Tris-HCl pH 7.5.
- Estimate the final DNA concentration and purity using spectrophotometry.
1B. Extraction of DNA (after in vitro infection of cultured cells)
- Maintain cultured Huh7-NTCP cells (11, 12) in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% v/v fetal bovine serum, 1 x Penicillin/Streptomycin, and 2 mM L-glutamine. This has previously been reported in detail (13).
- Seed Huh7-NTCP cells into a 12-well plate with 1 mL of DMEM containing 2x105 cells/mL.
- If testing cell treatments (e.g. HBV inhibitors), apply these to the culture supernatant 4 hours after seeding (1 day prior to HBV infection). For a negative control, use 200nM Myrcludex B (a potent HBV entry inhibitor (14)).
- Use heparin-column purified (15) supernatant from HepAD38 (16) as an inoculum to infect cells at 1000 VGE/cell in 500 µL of culture media (DMEM supplemented with 10% v/v fetal bovine serum, 1 x Penicillin/Streptomycin, 2 mM L-glutamine, and 2.5% v/v DMSO), containing 4% w/v polyethylene glycol 8000 (dissolved in 1x phosphate-buffered saline; PBS).
- Culture cells in a 37°C incubator (set at 5% CO2 and 90% humidity) overnight.
- Wash twice with 1 mL sterile 1x PBS at 16-24 hours post-infection.
- Replace culture media every two days following HBV infection until harvest.
- At day 3 post-infection, treat cells with 5 µM Tenofovir disoproxil and 10 µM Lamivudine to limit production of HBV replicative intermediates that are amplifiable by invPCR.
- At day 5 post-infection, trypsinise cells with 200 µL Trypsin-EDTA and resuspend them in 2 mL of DMEM (supplemented with 10% v/v fetal bovine serum, 1x Penicillin/Streptomycin, and 2mM L-glutamine), containing 5 µM Tenofovir disoproxil and 10 µM Lamivudine.
- Transfer the cell suspensions to a 6-well plate to induce one round of mitosis, which has been reported to induce loss of HBV cccDNA (17) and other HBV DNA intermediates that are amplifiable by invPCR.
- At day 7 post-infection, trypsinise the expanded cells in 400 µL Trypsin-EDTA and resuspend the mixture in 1 mL DMEM.
- Place the suspension in a 1.5 mL Eppendorf tube, pellet cells by centrifugation at 500 x g for 5 minutes, and remove supernatant by aspiration.
- (Optional) Store cell pellets at -20°C until ready for DNA extraction.
- Extract DNA from the cell pellet as above, or by using a column-based DNA extraction kit as per the manufacturer’s instructions.
- Estimate the final DNA concentration and purity using spectrophotometry. The DNA yield in a 100 μL elution volume is generally ~250-400 ng/μL.
- Inversion of DNA
- Aliquot ~1.5-2.5 μg of the total DNA extract into a 200 μL PCR tube.
- Add the appropriate amount of restriction enzyme master-mix to result in a 40μL reaction volume, containing 1x CutSmart buffer and 10U NcoI-HF.
- Mix reactions thoroughly and spin down in a small tube centrifuge.
- Incubate the restriction enzyme reaction in a PCR machine at 37°C for 1 hour for optimal digestion efficiency.
- Inactivate the restriction enzyme by incubating at 80°C for 20 minutes.
- Transfer the entire restriction enzyme reaction into a 1.5 mL Eppendorf tube.
- Add 400 μL of 1x T4 DNA ligase buffer and 500 U T4 DNA ligase and mix thoroughly. The large reaction volume encourages intra-molecular (as opposed to inter-molecular) ligation of the digested DNA fragments.
- Incubate the ligation reaction at room temperature for 2 hours to ensure complete ligation.
- Inactivate the T4 DNA ligase at 70°C for 20 minutes.
- Add 10 μL of 10% w/v sodium dodecyl sulphate to ensure complete inactivation of the T4 ligase.
- Mix the tubes by pulse vortexing and briefly spin down the reaction mix.
- Add NaCl to a final concentration of 100 mM and dextran (35 -45 kDa) to a final concentration of 90 µg/mL.
- Mix the tubes by pulse vortexing and briefly spin down the reaction mix.
- Add 900μL of 100% ethanol and mix by inversion.
- Precipitate the DNA at -20°C overnight.
- Pellet the precipitated DNA by centrifugation at 14,000 x g for 15 minutes.
- Wash the pellet with 500 μL of 70% v/v Ethanol and centrifugation at 14,000 x g for another 15 minutes.
- Remove the ethanol by aspiration with a P200 pipette.
- Air-dry the DNA pellet at RT for 20 minutes.
- Redissolve the pellet in 20 μL H2
- Add 20 μL of a restriction enzyme master-mix to result in a 40 μL reaction volume, containing 1x CutSmart buffer and 5U BsiHKAI and 5U SphI-HF.
- Incubate the restriction enzyme reaction in a heat-block at 37°C (the optimal temperature for SphI-HF) for 1 hr.
- Incubate the restriction enzyme reaction in a heat-block at 65°C (the optimal temperature for BsiHKAI) for 1 hr.
- Store the inverted DNA at -20°C until required.
- Nested PCR
- Prepare 1 mL of 1x Amplitaq Gold PCR mix containing the outer forward and reverse primers at a concentration of 0.5 µM.
- Add 170 μL of the 1x Amplitaq Gold PCR mix to wells A1 and E1 of a 96-well PCR plate.
- Add 120 μL of the 1x Amplitaq Gold PCR mix to wells B1 to H1.
- For cell culture samples, as significant clonal proliferation has not occurred, 2 different inverted samples can generally be analysed on the same PCR plate. In this case, add 10 μL of the inverted DNA to wells A1 and E1. For liver tissue samples, use only one DNA extract per plate and add inverted DNA to only well A1.
- Mix the reaction in each well by gently pipetting ~10 times using a P1000 set at 100 μL.
- Serially dilute samples from wells A1 to D1 (or A1 to H1 for tissue samples) at a ratio of 1:3 by transferring 60 μL at each step. Mix at each step by gently pipetting ~10 times using a P1000 set at 100 μL. Avoid forming bubbles.
- Repeat Step 3.3 for well E1, diluting down to well H1.
- Aliquot 10 μL of the reaction mixture using a multi-channel pipette from wells A1-H1 into wells A2-H2, A3-H3, and so forth until wells A12-H12 of the 96-well plate.
- Remove potential amplicons from a reusable silicon mat seal using DNAZap, rinse the mat thoroughly with DNA-free water and air-dry.
- Cover the PCR plate with the dry silicon mat, pressing firmly.
- Place the plate in a PCR machine and run the following program: 10 min at 95°C; 35 cycles of 15 sec at 95°C, 15 sec at 54°C and 3 min at 72°C; 7 min at 72°C, then hold at room temperature.
- Carefully remove the silicon mat to avoid cross-contamination between wells.
- Heat the pins of a 96-pin replicator to red-hot with a Bunsen burner and then cool for at least 5 minutes at room-temperature.
- Fill the wells of a second PCR plate with 10 μL of GoTaq Flexi Green PCR mix and the inner forward and reverse primers at 0.5 µM.
- Use the cooled replicator to transfer PCR products of the first 96-well plate to a second 96-well plate.
- Carry out the nested PCR using the same conditions as Step 3.10, except for changing the initial denaturation step at 95°C from 10 minutes to 2 minutes.
- PCR product isolation and gel extraction
- Analyse the PCR products by gel electrophoresis using a 96-well 1.3% w/v agarose gel. For a 100mL agarose gel, run at 200V for 10-15 minutes.
- Excise the DNA bands from agarose gels using disposable drinking straws. It is sufficient to isolate bands only from those dilutions where single PCR products can be resolved.
- For each PCR product, place the straw and agarose gel plug into a 1.5mL Eppendorf tube and then trim the straw to size with scissors.
- (Optional) Store at tubes -20°C for later extraction.
- Squeeze each straw to liberate the agarose plug into each Eppendorf tube.
- Add 300 μL of QX1 Buffer (Qiagen), 5 μL of QiaexII glass beads (Qiagen) to each tube.
- Extract the PCR products as per manufacturer’s instructions for the QiaexII gel extraction kit (except using half volumes for washing steps) and elute DNA from the beads with 30 μL of water or Elution Buffer.
- Submit purified DNA for Sanger sequencing using inner forward primer used in the nested PCR.
- Confirm virus-cell DNA junctions by nucleotide BLAST analysis (using default settings aligning to the entire nucleotide collection). Trim the 5’ HBV DNA sequence before re-running analysis if only partial alignment of the sequence is observed.
- Calculate the integration frequency by multiplying the dilution factor of inverted DNA templates by the number of virus-cell junctions detected at that dilution, followed by normalisation to the amount of total DNA input into the inversion reaction. Generally, the integration frequency is on the order of 1 in ~104
Table 1. PCR primers used in invPCR, reproduced from (6).
| PCR |
Forward Primer Sequence
(5’ -> 3’)+ |
Position on HBV genome* |
Reverse Primer Sequence
(5’ -> 3’)+ |
Position on HBV genome* |
| Outer |
TTCGCTTCACCTCTGCACG |
1603-1621 |
AAAGGACGTCCCGCGCAG |
1422-1405 |
| AAAGGACGTCCCGCGAAG |
1422-1405 |
| Inner |
TGGAGACCACCGTGAACG |
1626-1643 |
AGTACAGCCTAGCAGCCAT |
1388-1370 |
| CGCATGGAGACCACCGTGA# |
1623-1641 |
CACACCCTAGCAGCCATGG |
1390-1372 |
| CACAGCCTAGCAGCCATGG# |
1390-1372 |
| CGCATGGAAACCACCGTGA |
1623-1640 |
CACAGCCTAGCAGCCATGG |
1390-1372 |
+ These outer and inner PCR primer sets were previously used by us to amplify VCJ by nested PCR. A single forward primer could be used with one of multiple reverse primers. The specific primer set used for any given patient was based on their compatibility with the DNA sequence of the infecting HBV DNA strain.
* Primer positions were based on numbering of the HBV DNA nucleotide sequence from NCBI Reference Sequence: NC_003977.2.
# These primers are compatible with the Genotype D ayw strain (NCBI Reference Sequence: NC_003977.2) used in the majority of laboratory experiments.
Tips
- As this is a highly sensitive technique that amplifies down to single copies of DNA template and uses the same PCR primers for separate samples, limiting DNA contamination is a major issue. General strategies to limit PCR contamination include:
- Identify and establish 4 physically separate areas including (from “more dirty” to “less dirty”):
- Tissue DNA extraction area (carried out in a biosafety cabinet with a UV lamp for decontamination).
- DNA extraction (pre- and post-PCR), inversion and sequencing reaction set-up area;
- Template addition to PCR and flamed-pin transfer area (we have used PCR hoods with a decontaminating UV lamp with good results);
- Stock solution and PCR master-mix set-up area to be used to prepare fresh dedicated reagents for carrying out the inversion reaction, PCR, and DNA extractions. Make up buffers from new powdered stocks.
- Have separate lab gowns (which are changed often) for all 4 areas and change gloves when moving from “more dirty” to “less dirty” areas.
- Limit cross-current air flow within the lab, especially in the “template addition to PCR” area. Cross-contamination of wells at this point will lead to inaccurate quantification of virus cell junctions (VCJ). To locate an area of the lab with the least cross-currents, we hold up a piece of tissue and measure the amount of air movement by eye. PCR hoods can also be used to limit these currents. Use negative control wells in the 96-well PCR to test for cross-contamination.
- Decontaminate all work surfaces with 0.5% sodium hypochlorite solution or DNA-Zap. Wipe off with a damp paper towel, and then dry with a clean paper towel.
- Never decontaminate with 70% ethanol for the purpose of removing DNA contaminants. Ethanol will fix any DNA to the surface, making it even harder to remove.
- To design a new inversion protocol, our suggested criteria for invPCR designs are as follows:
- As a guideline restriction enzyme 1, 2 and 3 sites (see Figure 1) should occur within ~1 kbp of the expected VCJ site on the HBV genome at ~nt 1832 (1, 18) to limit the size, and so improve circularisation and PCR amplification of the excised fragments.
- The design must allow >40 nt between the RE2 restriction site and the expected VCJ site to allow nested primer design.
- Similarly, the design must allow >40 nt between the RE1 and RE2 sites on the HBV genome for nested primer design.
- RE1 site should occur as frequently as possible in the host genome, but occur very rarely (preferably uniquely) in the HBV DNA genome to limit formation of amplifiable cccDNA-derived fragments.
- RE1 should be able to be heat-inactivated to simplify transition to the ligation step.
- RE2 and particularly RE3 should occur as frequently as possible in the HBV genome to limit formation of amplifiable cccDNA-derived fragments, but occur rarely in the human genome to avoid cleavage between the VCJ and a downstream RE1 site, resulting in an unamplifiable fragment.
- To keep the liver tissue cool during sectioning prior to DNA extraction, we place an inverted plastic petri dish or lid on top of dry ice. We then put the frozen tissue in the inverted lid and break into 5 mg fragments (we generally analyse five 5 mg liver fragments per patient), changing dishes and scalpels for each patient. If forceps are used during the extraction procedure, immerse in 0.5% sodium hypochlorite solution for at least 15 min between samples, rinse in water and dry on paper towels.
- Using the inversion and PCR amplification conditions described, full-length cccDNA should not amplify to detectable levels. However, it may still be necessary to purify high molecular weight DNA to reduce cccDNA contamination, since residual amplification may result due to 1) naturally occurring deletion mutants of cccDNA, or 2) low level star activity of RE1 and circularisation of cccDNA fragments. In each case, the DNA molecules generated are small enough to be amplified to detectable levels. An additional gel purification step using 1% low-melting temperature agarose (Bio-Rad) can be done before the inversion step to isolate high-molecular weight cellular DNA from low (<3.2 kb) molecular weight intermediates. It is important to quantify the DNA before and after this step to account for any lost DNA yield during the purification in order to calculate VCJ number in the original DNA sample.
- As a lower concentration of DNA should improve the inversion efficiency, less DNA can be put into the assay. The amount of DNA can be determined by probability calculations of DNA fragment self-ligation as described below.
- The large 400 μL ligation reaction volume favours self-ligation and circularisation of DNA fragments as opposed to intramolecular ligation. The probability of circularization of linear DNA during ligation is governed by the ratio of factors j and i (19),where j is the effective concentration of one end of a DNA molecule in the neighbourhood of the other end of the same molecule in ends/mL and i is the total concentration of DNA ends within a given solution in ends/mL. The probability of circularisation can be shown as simply j/i. When j/i = 1, equal products of circular and linear ligations are expected. When j/i > 1, self-ligated circular forms are favoured and when j/i < 1, intermolecular linear ligation is preferred. Covalently closed circular structures of DNA can be found only when j/i < 2-3 (19). Using the assumptions that:
- RE1-cleaved VCJ-containing fragments are 1000 bp long;
- 75 μg of DNA in the ligation reaction (over-estimate);
- Cellular DNA has a random distribution of bases ;
- RE1 has a 4 base recognition sequence; and
- 400 uL ligation volume,
We calculated a
j/
i ratio of 3.84. Therefore, with the above assumptions, circularisation of DNA fragments is preferred over inter-molecular ligation during the inversion reaction.
- In some instances when DNA concentrations are low (e.g. extracts from laser-microdissected tissue samples), we do not do the serial dilution step; instead we add the inverted DNA mixture directly to 1 mL of Amplitaq Gold PCR mix, vortex briefly and distribute 10 μL of PCR mix to each well, after which we proceed straight to the PCR step.
- Serial dilutions should be mixed by slowly pipetting up and down 10 times using a 1.0 mL pipette set at ~100 μL. This will limit bubbles and cross-contamination of PCR products.
- Heat the 96-pin replicator until the pins are red hot. Note that the heated 96-pin replicator will cause convection air currents, so cool for 5-10 min at RT before transferring the templates from the first PCR tray to the second. The replicator will transfer only ~1 μL. Some carbon flecks may also be transferred, but are sterile and should not affect the PCR.